The Smoking Bat: The Quest To Uncover the Origins of SARS-CoV-2
The World Health Organization’s recent probe into the origins of COVID-19 in Wuhan has renewed interest in piecing together when, where, and how the virus jumped from animals to humans during the fall of 2019. The Wuhan Seafood Market outbreak appears to be a red herring, serving as an early amplifier of a simmering outbreak in December 2019, but not the original source of the animal virus, which jumped to humans weeks, or possibly months, earlier (1). The pangolin’s brief moment of fame also appears to have been a red herring, as the pangolin viruses were only distantly related to the human viruses (2). Then again, we know previous little about coronavirus diversity in pangolins and other species. As attention focuses on the first human cases in China, including the unreleased raw data, there has been a notable lack of public interest in what the virus was doing before its grand entrance in humans and how little coronavirus data is available from animals.

Photo by Warner Bros/Kobal/Shutterstock Kate Winslet Contagion – 2011
Contagion got many things right about outbreak investigations, but the film oversimplified how scientists trace a pandemic’s origins back to animals. At no time in history has an outbreak’s zoonotic source been solved using security camera footage. Anyone who has searched for the animal origins of Ebola outbreaks in bats in West Africa knows how painstakingly laborious the task of sampling wildlife is. Theoretically livestock are easier because at least we know where they are housed. But the politics of getting access to samples can be equally daunting when a wrong bug found in a herd could cost a farmer their lifesavings.

Did SARS-CoV-2 come from traded wildlife, such as pangolins?
But as humans grapple with the current COVID-19 crisis, the ancestor of the next pandemic virus is already lurking in an animal host somewhere. It could be in a bat in China, a primate in sub-Saharan Africa, or a pig in the Americas. The virus could be a decade away from acquiring the properties needed to transmit to humans, or it could be weeks. Despite the disruptions caused by COVID-19, humans continue to create conditions for zoonotic transfer of viruses from animals to humans, consuming millions of animals every day, congregating in live animal markets, and even holding pig shows. Imagine another pandemic on top of COVID-19.
How can we learn more about a zoonotic event that occurred over a year ago? For approximately the same price tag as what the UK government is currently investing to characterize several thousand SARS-CoV-2 viruses a week (3), researchers could at least begin a sweeping survey of the genetic composition of hundreds of thousands of coronaviruses circulating in wildlife, livestock, and live animal markets across east Asia. A region-wide survey conducted a year following the original animal-to-human transmission event is not guaranteed to produce the ‘smoking gun’ virus or pinpoint which market or farm was responsible. But the descendants of the virus that jumped to humans continue to circulate in animals in China and other Asian countries, and the genetic material of each virus provides important clues about its grandparents. Scientists can reconstruct decades of evolutionary history from gene sequence alone.
Mexico’s response to the 2009 H1N1 influenza pandemic proves how decoding viruses in animals even years after the event can resolve the mysteries of a pandemic’s origins and provide the global community clues for how to prevent the next one. At first, scientists thought the virus that caused the global pandemic in 2009 came from pigs in China, where the most closely related animal flu viruses were found (4). China seemed a logical source, given that the last two influenza pandemics originated in China and the country is home to half of the world’s pigs. Although the first signs of the virus transmitting in humans occurred in Mexico (5), Mexican pigs seemed like an unlikely source because the virus contained genetic material that had never been seen in pigs in any country in the Americas. For seven years it remained a mystery how a virus that seemed to come from Chinese pigs presumably sparked its first human outbreak in Mexico. Had Chinese pigs been smuggled into Mexico? Or had Chinese pig farmers vacationed in Cancun just after acquiring the virus?
To its credit, Mexico resolved the mystery by embarking after the pandemic on new surveillance of influenza in its swine herds scattered across the northern, central, and far eastern regions of the country. By working with Mexican swine veterinarians our research team uncovered a surprising find: viruses similar in important ways to those in Chinese pigs (6). The story told how Mexico’s rapid modernization of swine production in the 1990s allowed viruses from different continents to be imported and hybridize with each other to create a pandemic in a country assumed to be low risk. While swine farmers care a great deal about keeping deadly pathogens like African swine fever from crossing borders, influenza is already in pigs in every country and newly imported strains are not perceived as a problem worthy of testing or quarantine from an animal health perspective.
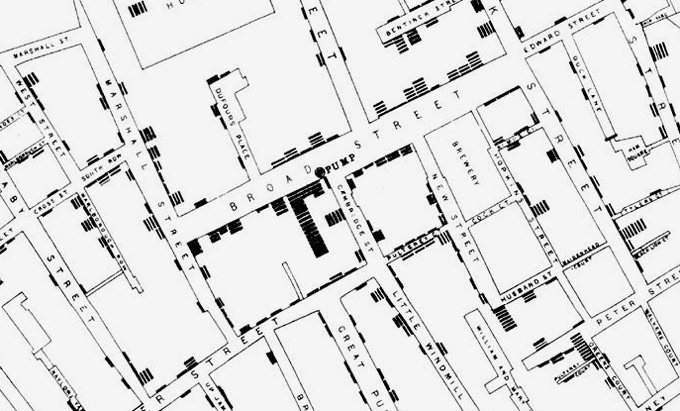
British physician John Snow’s 1854 map of cholera cases in Soho, London
Altering food production is economically costly and socially disruptive. Just as COVID-19 lockdowns have become less blunt as key vectors of community transmission were identified, changing how we operate live animal markets, wild animal trade, or bushmeat needs to be grounded in data. Two centuries ago, the grandfather of modern epidemiology John Snow was successful because he used rigorously collected data to pinpoint a single contaminated water pump that was causing the bulk of cholera deaths, and did not ask authorities to disengage every pump in the city. But scientists need a great deal more data to track disease transmission in animals at anything approaching the levels we achieve in humans. We can uncover how coronaviruses get shuttled between countries and continents by trade of wildlife and livestock, as well as by movements of wildlife displaced by habitat loss, and how this relates to human coronavirus outbreaks. Although viruses similar to SARS-CoV-1 in 2003 were identified in civet cats in a live animal market in China (7), we do not know how those viruses wound up in civets in the first place. Are pangolins or other traded wildlife species an important permanent reservoir for coronaviruses connected to human outbreaks? Where did the four coronaviruses that cause common colds in humans come from? Again, the limiting factor is data.
With over two million COVID-19 deaths it may seem pointless to figure out where the virus came from originally. But fundamental insights into disease systems can have impacts far beyond a single disease event. Although John Snow only asked authorities to turn off a single water pump, the insight that cholera epidemics might be vanquished by cleaning up water supplies transformed the 19th century. The construction of modern sewer systems turned the Industrial Revolution’s squalid, disease-ridden cities in Europe and America into modern economic centers capable of supporting skyrocketing densities. Some day we may back look at the dark ages of 2020 with the same curiosity as ‘night soil men’ pushing carts of human feces and horse dung down Broadway.

Today we associate sewers with rats and odors, but at the time they were marvels of engineering.
TL;DR. Half the world thinks SARS-CoV-2 came from a lab escape. The other half thinks someone ate a bat. Scientists can do a great deal better, and avert future pandemics, but they need data from animals.
References
- Pekar J, Worobey M, Moshiri N, Scheffler K, Wertheim JO. Timing the SARS-CoV-2 Index Case in Hubei Province. bioRxiv [Preprint]. 2020 Nov 24:2020.11.20.392126.
- Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC, Tong YG, Shi YX, Ni XB, Liao YS, Li WJ, Jiang BG, Wei W, Yuan TT, Zheng K, Cui XM, Li J, Pei GQ, Qiang X, Cheung WY, Li LF, Sun FF, Qin S, Huang JC, Leung GM, Holmes EC, Hu YL, Guan Y, Cao WC. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020 Jul;583(7815):282-285.
- Nelson MI. 2021. Tracking the UK SARS-CoV-2 outbreak. Science. 371(6530):680-681.
- Grantz KH, Meredith HR, Cummings DAT, Metcalf CJE, Grenfell BT, Giles JR, Mehta S, Solomon S, Labrique A, Kishore N, Buckee CO, Wesolowski A. The use of mobile phone data to inform analysis of COVID-19 pandemic epidemiology. Nat Commun. 2020 Sep 30;11(1):4961.
- Chowell G, Bertozzi SM, Colchero MA, Lopez-Gatell H, Alpuche-Aranda C, Hernandez M, Miller MA. 2009. Severe respiratory disease concurrent with the circulation of H1N1 influenza. N Engl J Med. 361(7):674-9.
- Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 459 (7250): 1122-5.
- Mena I, Nelson MI, Quezada-Monroy F, Dutta J, Cortes-Fernández R, Lara-Puente JH, Castro-Peralta F, Cunha L, Trovao NS, Lozano-Dubernard B, Rambaut A, van Bakel H, García-Sastre A. 2016. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. eLife 5: e16777.
- Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JS, Poon LL. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003 Oct 10;302(5643):276-8.
- Chen Y, Trovão NS, Wang G, Zhao W, He P, Zhou H, Mo Y, Wei Z, Ouyang K, Huang W, García-Sastre A, Nelson MI. Emergence and Evolution of Novel Reassortant Influenza A Viruses in Canines in Southern China. mBio. 2018 Jun 5;9(3):e00909-18

Leave a Reply
Want to join the discussion?Feel free to contribute!